7 Internal Loops
7.1 1×1, 1×2, 2×2 Internal Loops
Small symmetric internal loops have tabulated free energy and enthalpy changes, where experimentally determined values are used if available.
7.2 Other Internal Loops
The stabilities of other internal loops are predicted using the equation:
ΔG°37 internal = ΔG°37 initiation(n) + ΔG°37 asymmetry × |n1 – n2| + ΔG°37 mismatch(mismatch 1) + ΔG°37 mismatch(mismatch 2) + ΔG°37 AU/GU closure(per AU or GU closure)
where the initiation is a length dependent term for n unpaired nucleotides, an asymmetry term is multiplied by the absolute value of the difference in the number of unpaired nucleotides on each side of the loop, and sequence-dependent mismatch terms are applied for first mismatches of specific sequences. The AU/GU closure is applied per AU or GU closing pair and is used instead of the AU or GU penalty at the end of the helix (see Watson-Crick-Franklin or GU pairs).
Experimental data for ΔG°37 initiation(n) is available for loops up to n = 6. For larger internal loops, an extrapolation is made:
ΔG°37 initiation(n>6) = ΔG°37 initiation(6) + 1.08 × ln(n/6)
Similarly, the enthalpy change is predicted with the equation:
ΔH°internal = ΔH°initiation(n) + ΔH°asymmetry × |n1 – n2| + ΔH°mismatch(mismatch 1) + ΔH°mismatch(mismatch 2) + ΔH°AU/GU closure(per AU or GU closure)
where terms are analagous to those for predicting folding free energy changes.
The mismatch parameters are sequence-dependent and are different for 1×(n-1) loop, 2×3 loop, and other internal loops. In the case of 1×(n–1) loops, the mismatches are set to 0 kcal/mol for free energy and enthalpy changes.
In the absence of data for loops larger than n=6, ΔH°initiation(n>6) = ΔH°initiation(6)
7.3 Examples
2×2 internal loop
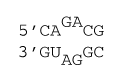
ΔG°37 = ΔG°37(Watson-Crick-Franklin stacks) + ΔG°37 intermolecular initiation + ΔG°37(Internal Loop)
ΔG°37 = ΔG°37(CG followed by AU) + ΔG°37(CG followed by GC) + ΔG°37 intermolecular initiation + ΔG°37(2×2 Internal Loop)
ΔG°37 = –2.11 kcal/mol – 2.36 kcal/mol +4.09 kcal/mol – 1.1 kcal/mol
ΔG°37 = –1.5 kcal/mol
ΔH° = ΔH°(Watson-Crick-Franklin stacks) + ΔH°intermolecular initiation + ΔH°(Internal Loop)
ΔH °= ΔH°(CG followed by AU) + ΔH°(CG followed by GC) + ΔH°intermolecular initiation+ ΔH°(2×2 Internal Loop)
ΔH° = –10.44 kcal/mol – 10.64 kcal/mol + 3.61 kcal/mol – 19.7 kcal/mol
ΔH° = –37.2 kcal/mol
Note that the internal loop lookup tables account for terminal AU pairs that are adjacent to internal loops.
1×5 internal loop
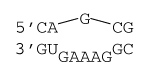
ΔG°37 = ΔG°37(Watson-Crick-Franklin stacks) + ΔG°37 intermolecular initiation + ΔG°37(Internal Loop)
ΔG°37 = ΔG°37(CG followed by AU) + ΔG°37(CG followed by GC) + ΔG°37 intermolecular initiation+ ΔG°37 initiation(6) + ΔG°37 asymmetry×|n1 - n2| + ΔG°37 mismatch(mismatch 1) + ΔG°37 mismatch(mismatch 2) + ΔG°37 AU/GU closure
ΔG°37 = –2.11 kcal/mol – 2.36 kcal/mol + 4.09 kcal/mol + 2.0 kcal/mol + 0.6×|1 - 5| kcal/mol + 0 kcal/mol + 0 kcal/mol + 0.7 kcal/mol
ΔG°37 = +4.7 kcal/mol
ΔH° = ΔH°(Watson-Crick-Franklin stacks) + ΔH°intermolecular initiation + ΔH°(Internal Loop)
ΔH°= ΔH°(CG followed by AU) + ΔH°(CG followed by GC) + ΔH°intermolecular initiation+ ΔH°initiation(6) + ΔH°asymmetry × |n1 - n2| + ΔH°mismatch(mismatch 1) + ΔH°mismatch(mismatch 2) + ΔH°AU/GU closure
ΔH° = –10.44 kcal/mol – 10.64 kcal/mol + 3.61 kcal/mol – 1.3 kcal/mol + 3.2×|1 - 5| kcal/mol + 0 kcal/mol + 0 kcal/mol + 5.0 kcal/mol
ΔH° = –1.0 kcal/mol
Note that the free energy and enthalpy changes for first mismatches in 1×(n-1) internal loops are 0 kcal/mol.
2×3 internal loop with stabilizing mismatches
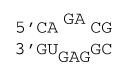
ΔG°37 = ΔG°37(Watson-Crick-Franklin stacks) + ΔG°37 intermolecular initiation + ΔG°37(Internal Loop)
ΔG°37 = ΔG°37(CG followed by AU) + ΔG°37(CG followed by GC) + ΔG°37 intermolecular initiation + ΔG°37 initiation(5) + ΔG°37 asymmetry×|n1 - n2| + ΔG°37 mismatch(mismatch 1) + ΔG°37 mismatch(mismatch 2) + ΔG°37 AU/GU closure
ΔG°37 = –2.11 kcal/mol – 2.36 kcal/mol + 4.09 kcal/mol + 2.0 kcal/mol + 0.6×|2 - 3| kcal/mol – 0.8 kcal/mol – 1.2 kcal/mol + 0.7 kcal/mol
ΔG°37 = +0.9 kcal/mol
ΔH° = ΔH°(Watson-Crick-Franklin stacks) + ΔH°intermolecular initiation + ΔH°(Internal Loop)
ΔH°= ΔH°(CG followed by AU) + ΔH°(CG followed by GC) + ΔH°intermolecular initiation + ΔH°initiation(5) + ΔH°asymmetry × |n1 - n2| + ΔH°mismatch(mismatch 1) + ΔH°mismatch(mismatch 2) + ΔH°AU/GU closure
ΔH° = –10.44 kcal/mol – 10.64 kcal/mol + 3.61 kcal/mol – 6.8 kcal/mol + 3.2×|2 - 3| kcal/mol – 9.0 kcal/mol – 10.9 kcal/mol + 5.0 kcal/mol
ΔH° = –36.0 kcal/mol
7.4 Parameter Tables
1×1 internal loop free energy change tables are available in text and html format. Enthalpy change tables are available in text and html format. Note that these tables incorporate the AU/GU closure penalties and therefore no AU/GU helix end penalty should be applied for internal loop closure.
1×2 internal loop free energy change tables are available in text and html format. Enthalpy change tables are available in text and html format. Note that these tables incorporate the AU/GU closure penalties and therefore no AU/GU helix end penalty should be applied for internal loop closure.
2×2 internal loop free energy change tables are available in text and html format. Enthalpy change tables are available in text and html format. Note that these tables incorporate the AU/GU closure penalties and therefore no AU/GU helix end penalty should be applied for internal loop closure.
Other parameters in text free energy or enthalpy or html format.
7.5 References
The internal loop nearest neighbor parameters for free energy change were reported in:
Mathews, D.H., Disney, M.D., Childs, J.L., Schroeder, S.J., Zuker, M. and Turner, D.H. (2004) Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. USA, 101, 7287-7292.
The enthalpy change parameters were reported in:
Lu, Z.J., Turner, D.H. and Mathews, D.H. (2006) A set of nearest neighbor parameters for predicting the enthalpy change of RNA secondary structure formation. Nucleic Acids Res., 34 4912 - 4924.
The experimental data for the fit of the parameters were taken from:
- Peritz, A.E., Kierzek, R., Sugimoto, N. and Turner, D.H. (1991) Thermodynamic study of internal loops in oligoribonucleotides: Symmetric loops are more stable than asymmetric loops. Biochemistry, 30, 6428-6436.
- SantaLucia, J., Jr., Kierzek, R. and Turner, D.H. (1991) Functional group substitutions as probes of hydrogen bonding between GA mismatches in RNA internal loops. J. Am. Chem. Soc., 113, 4313-4322.
- SantaLucia, J., Jr., Kierzek, R. and Turner, D.H. (1991) Stabilities of consecutive A.C, C.C, G.G, U.C, and U.U mismatches in RNA internal loops: evidence for stable hydrogen-bonded U.U and C.C+ pairs. Biochemistry, 30, 8242-8251.
- Walter, A.E., Wu, M. and Turner, D.H. (1994) The stability and structure of tandem GA mismatches in RNA depend on closing base pairs. Biochemistry, 33, 11349-11354.
- Wu, M., McDowell, J.A. and Turner, D.H. (1995) A periodic table of symmetric tandem mismatches in RNA. Biochemistry, 34, 3204-3211.
- Schroeder, S., Kim, J. and Turner, D.H. (1996) G.A and U.U mismatches can stabilize RNA internal loops of three nucleotides. Biochemistry, 35, 16105-16109.
- Xia, T., McDowell, J.A. and Turner, D.H. (1997) Thermodynamics of nonsymmetric tandem mismatches adjacent to G.C base pairs in RNA. Biochemistry, 36, 12486-12487.
- Kierzek, R., Burkard, M. and Turner, D. (1999) Thermodynamics of single mismatches in RNA duplexes. Biochemistry, 38, 14214-14223.
- Schroeder, S.J. and Turner, D.H. (2000) Factors affecting the thermodynamic stability of small asymmetric internal loops in RNA. Biochemistry, 39, 9257-9274.
- Burkard, M.E., Xia, T. and Turner, D.H. (2001) Thermodynamics of RNA internal loops with a guanosine-guanosine pair adjacent to another noncanonical pair. Biochemistry, 40, 2478-2483.
- Schroeder, S.J. and Turner, D.H. (2001) Thermodynamic stabilities of internal loops with GU closing pairs in RNA. Biochemistry, 40, 11509-11517.
- Schroeder, S.J., Fountain, M.A., Kennedy, S.D., Lukavsky, P.J., Puglisi, J.D., Krugh, T.R. and Turner, D.H. (2003) Thermodynamic stability and structural features of the J4/5 loop in a Pneumocystis carinii group I intron. Biochemistry, 42, 14184-14196.
