3 Dangling Ends
Dangling ends are nucleotides that stack on the ends of helices. In secondary structures, they occur in multibranch and exterior loops. They occur as either \(5'\) dangling ends (an unpaired nucleotide \(5'\) to the helix end) or \(3'\) dangling ends (an unpaired nucleotide \(3'\) to the helix end). In RNA, \(3'\) dangling ends are generally more stabilizing than \(5'\) dangling ends. Note that if a helix end is extended on both the \(5'\) and \(3'\) strands, then a terminal mismatch exists (not the sum of \(5'\) and \(3'\) dangling ends).
3.1 Example
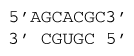
ΔG°37 = ΔG°37(Watson-Crick-Franklin Helix) + ΔG°37(\(3'\) dangling C adjacent to GC) + ΔG°37(\(5'\) dangling A adjacent GC)
ΔG°37 = –6.04 kcal/mol – 0.4 kcal/mol – 0.2 kcal/mol
ΔG°37 = –6.6 kcal/mol
ΔH° = ΔH°(Watson-Crick-Franklin Helix) + ΔH°(\(3'\) dangling C adjacent to GC) + ΔH°(\(5'\) dangling A adjacent GC)
ΔH° = –43.75 kcal/mol – 2.8 kcal/mol – 1.6 kcal/mol
ΔH° = –48.2 kcal/mol
Note that this example contains both a \(5'\) and a \(3'\) dangling end (at opposite ends of the duplex).
3.2 Tables
The tables of parameters are available as plain text for free energy change, plain text for enthalpy change, or html.
3.3 References
The dangling end parameters were assembled in:
Serra, M.J. and Turner, D.H. (1995) Predicting Thermodynamic Properties of RNA. Methods Enzymol., 259, 242-261.
The optical melting experiments for dangling ends were reported in:
- Freier, S.M., Burger, B.J., Alkema, D., Neilson, T. and Turner, D.H. (1983) Effects of \(3'\) dangling end stacking on the stability of GGCC and CCGG double helices. Biochemistry, 22, 6198-6206.
- Petersheim, M. and Turner, D.H. (1983) Base-stacking and base-pairing contributions to helix stability: thermodynamics of double-helix formation with CCGG, CCGGp, CCGGAp, ACCGGp, CCGGUp, and ACCGGUp. Biochemistry, 22, 256-263.
- Freier, S.M., Alkema, D., Sinclair, A., Neilson, T. and Turner, D.H. (1985) Contributions of dangling end stacking and terminal base-pair formation to the stabilities of XGGCCp, XCCGGp, XGGCCYp, and XCCGGYp helixes. Biochemistry, 24, 4533-4539.
- Freier, S.M., Kierzek, R., Caruthers, M.H., Neilson, T. and Turner, D.H. (1986) Free energy contributions of G.U and other terminal mismatches to helix stability. Biochemistry, 25, 3209-3223.
- Freier, S.M., Sugimoto, N., Sinclair, A., Alkema, D., Neilson, T., Kierzek, R., Caruthers, M.H. and Turner, D.H. (1986) Stability of XGCGCp, GCGCYp, and XGCGCYp helixes: an empirical estimate of the energetics of hydrogen bonds in nucleic acids. Biochemistry, 25, 3214-3219.
- Sugimoto, N., Kierzek, R. and Turner, D.H. (1987) Sequence dependence for the energetics of dangling ends and terminal base pairs in ribonucleic acid. Biochemistry, 26, 4554-4558.
- Turner, D.H., Sugimoto, N. and Freier, S.M. (1988) RNA structure prediction. Ann. Rev. Biophys. Biophys. Chem., 17, 167-192.
- Longfellow, C.E., Kierzek, R. and Turner, D.H. (1990) Thermodynamic and spectroscopic study of bulge loops in oligoribonucleotides. Biochemistry, 29, 278-285.
