11 Watson-Crick-Franklin Helices
11.1 Free Energy Change at 37 °C
Folding free energy changes for Watson-Crick-Franklin helices are predicted using the equation:
ΔG°37 Watson-Crick-Franklin = ΔG°37 intermolecular initiation + ΔG°37 AU end penalty (per AU end) + ΔG°37 symmetry (self-complementary duplexes) + Σ[ΔG°37 stacking]
where intermolecular initiation is applied for bimolecular structure formation, the AU end penalty is applied once per each AU pair at the end of a helix, the symmetry correction is applied to self-complementary duplexes, and the stacking term is a sum of sequence-dependent parameters over all base pair stacks. For helices of P uninterrupted basepairs, there are P-1 stacks of pairs.
11.2 Examples
11.2.1 Self complementary duplex
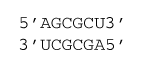
ΔG°37 = ΔG°37 intermolecular initiation + 2×ΔG°37 AU end penalty+ ΔG°37 symmetry + ΔG°37(AU followed by GC) + ΔG°37(GC followed by CG) + ΔG°37(CG followed by GC) + ΔG°37(GC followed by CG) + ΔG°37(CG followed by UA)
ΔG°37 = 4.09 kcal/mol + 2×0.45 kcal/mol + 0.43 kcal/mol –2.08 kcal/mol – 3.42 kcal/mol – 2.36 kcal/mol – 3.42 kcal/mol – 2.08 kcal/mol
ΔG°37 = –7.94 kcal/mol
Note that, for example, the parameter for (AU followed by GC) is the same as (CG followed by UA) because the correct directionality of the strands is preserved.
Non-self complementary duplex
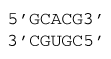
ΔG°37 = ΔG°37 intermolecular initiation + ΔG°37(GC followed by CG) + ΔG°37(CG followed by AU) + ΔG°37(AU followed by CG) + ΔG°37(CG followed by GC)
ΔG°37 = 4.09 kcal/mol – 3.42 kcal/mol – 2.11 kcal/mol – 2.24 kcal/mol – 2.36 kcal/mol
ΔG°37 = –6.04 kcal/mol
11.3 Tables
The table of parameters is available as plain text (including GU pairs; see the GU section for special cases for 5’GGUC/3’CUGG and 5’ GG/3’UU motifs) or html.
11.4 References
The Watson-Crick-Franklin nearest neighbor parameters were reported in:
Xia, T., SantaLucia, J., Jr., Burkard, M.E., Kierzek, R., Schroeder, S.J., Jiao, X., Cox, C. and Turner, D.H. (1998) Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick pairs. Biochemistry, 37, 14719-14735.
The experimental data for the fit of the parameters were taken from:
- Nelson, J.W., Martin, F.H. and Tinoco, I., Jr. (1981) DNA and RNA oligomer thermodynamics: the effect of mismatched bases on double-helix stability. Biopolymers, 20, 2509-2531.
- Freier, S.M., Burger, B.J., Alkema, D., Neilson, T. and Turner, D.H. (1983) Effects of 3’ dangling end stacking on the stability of GGCC and CCGG double helices. Biochemistry, 22, 6198-6206.
- Petersheim, M. and Turner, D.H. (1983) Base-stacking and base-pairing contributions to helix stability: thermodynamics of double-helix formation with CCGG, CCGGp, CCGGAp, ACCGGp, CCGGUp, and ACCGGUp. Biochemistry, 22, 256-263.
- Freier, S.M., Alkema, D., Sinclair, A., Neilson, T. and Turner, D.H. (1985) Contributions of dangling end stacking and terminal base-pair formation to the stabilities of XGGCCp, XCCGGp, XGGCCYp, and XCCGGYp helixes. Biochemistry, 24, 4533-4539.
- Freier, S.M., Sinclair, A., Neilson, T. and Turner, D.H. (1985) Improved free energies for G-C base-pairs. J. Mol. Biol., 185, 645-647.
- Hickey, D.R. and Turner, D.H. (1985) Solvent effects on the stability of A7U7p. Biochemistry, 24, 2086-2094.
- Freier, S.M., Kierzek, R., Caruthers, M.H., Neilson, T. and Turner, D.H. (1986) Free energy contributions of G.U and other terminal mismatches to helix stability. Biochemistry, 25, 3209-3223.
- Freier, S.M., Kierzek, R., Jaeger, J.A., Sugimoto, N., Caruthers, M.H., Neilson, T. and Turner, D.H. (1986) Improved free-energy parameters for predictions of RNA duplex stability. Proc. Natl. Acad. Sci. USA., 83, 9373-9377.
- Kierzek, R., Caruthers, M.H., Longfellow, C.E., Swinton, D., Turner, D.H. and Freier, S.M. (1986) Polymer-supported synthesis and its application to test the nearest-neighbor model for duplex stability. Biochemistry, 25, 7840-7846.
- Sugimoto, N., Kierzek, R., Freier, S.M. and Turner, D.H. (1986) Energetics of internal GU mismatches in ribooligonucleotide helixes. Biochemistry, 25, 5755-5759.
- Sugimoto, N., Kierzek, R. and Turner, D.H. (1987) Sequence dependence for the energetics of dangling ends and terminal base pairs in ribonucleic acid. Biochemistry, 26, 4554-4558.
- Longfellow, C.E., Kierzek, R. and Turner, D.H. (1990) Thermodynamic and spectroscopic study of bulge loops in oligoribonucleotides. Biochemistry, 29, 278-285.
- Hall, K.B. and McLaughlin, L.W. (1991) Thermodynamic and structural properties of pentamer DNA.DNA, RNA.RNA, and DNA.RNA duplexes of identical sequence. Biochemistry, 30, 10606-10613.
- He, L., Kierzek, R., SantaLucia, J., Jr., Walter, A.E. and Turner, D.H. (1991) Nearest-neighbor parameters for G.U mismatches. Biochemistry, 30, 11124-11132.
- Peritz, A.E., Kierzek, R., Sugimoto, N. and Turner, D.H. (1991) Thermodynamic study of internal loops in oligoribonucleotides: Symmetric loops are more stable than asymmetric loops. Biochemistry, 30, 6428-6436.
- Walter, A.E., Wu, M. and Turner, D.H. (1994) The stability and structure of tandem GA mismatches in RNA depend on closing base pairs. Biochemistry, 33, 11349-11354.
- Wu, M., McDowell, J.A. and Turner, D.H. (1995) A periodic table of symmetric tandem mismatches in RNA. Biochemistry, 34, 3204-3211.
- McDowell, J.A., He, L., Chen, X. and Turner, D.H. (1997) Investigation of the structural basis for thermodynamic stabilities of tandem GU wobble pairs: NMR structures of (rGGAGUUCC)2 and (rGGAUGUCC)2. Biochemistry, 36, 8030-8038.
- Xia, T., McDowell, J.A. and Turner, D.H. (1997) Thermodynamics of nonsymmetric tandem mismatches adjacent to G.C base pairs in RNA. Biochemistry, 36, 12486-12487.
- Xia, T., SantaLucia, J., Jr., Burkard, M.E., Kierzek, R., Schroeder, S.J., Jiao, X., Cox, C. and Turner, D.H. (1998) Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick pairs. Biochemistry, 37, 14719-14735.
