5 Hairpin Loops
5.1 Folding Free Energy Change
Hairpin loops of 4 or more nucleotides
The prediction of folding free energy changes for hairpins of 4 or more unpaired nucleotides is made with the following equation (also see Examples below):
ΔG°37 hairpin (>3 nucleotides in loop) = ΔG°37 initiation (n) + ΔG°37 (terminal mismatch) + ΔG°37 (UU or GA first mismatch) + ΔG°37 (GG first mismatch) + ΔG°37 (special GU closure) + ΔG°37 penalty (all C loops)
In this equation, n is the number of nucleotides in loop, the terminal mismatch parameter is the sequence-dependent term for the first mismatch stacking on the terminal base pair, UU and GA first mismatches receive a bonus (not applied to AG first mismatches), GG first mismatches receive a bonus, the special GU closure term is applied only to hairpins in which a GU closing pair (not UG) is preceded by two Gs, and finally loops with all C nucleotides receive a penalty.
The penalty for all C loops longer than C3 is:
ΔG°37 penalty (all C loops with n > 3) = An + B
Hairpin loops of 3 unpaired nucleotides
For hairpin loops of 3 nucleotides, the folding free energy change is estimated using:
ΔG°37 hairpin (3 unpaired nucleotides) = ΔG°37 initiation (3) + ΔG°37 penalty (C3 loop)
As opposed to longer hairpin loops, hairpin loops of three nucleotides do not receive a sequence-dependent first mismatch term. All C hairpin loops of three nucleotides receive a stability penalty.
Special hairpin loops
There are hairpin loop sequences of 3, 4, and 6 nucleotides that have stabilities poorly fit by the model. These hairpins are assigned stabilities based on experimental data.
Short hairpin loops
The nearest neighbor rules prohibit hairpin loops with fewer than 3 nucleotides.
5.2 Folding Enthalpy Change
Hairpin loops of 4 or more nucleotides
The prediction of folding enathlpy changes for hairpins of 4 or more nucleotides is made with the following equation:
ΔH°hairpin (>3 unpaired nucleotides) = ΔH°initiation (n) + ΔH° (terminal mismatch) + ΔH° (UU or GA first mismatch) + ΔH° (special GU closure) + ΔH° (all C loops)
As with the free energy change equation above, n is the number of nucleotides in the loop, the terminal mismatch parameter is the sequence-dependent term for the first mismatch stacking on the terminal pair, UU and GA first mismatches receive a bonus (not applied to AG first mismatches), the special GU closure term is applied only to hairpins in which a GU closing pair (not UG) is preceded by two Gs, and finally the all C loops receive a penalty.
The penalty for all C loops longer than C3 is:
ΔH°penalty (all C loops with n > 3) = A’n + B’
Hairpin loops of 3 nucleotides
For hairpin loops of 3 unpaired nucleotides, the enthalpy change is estimated using:
ΔH°hairpin (3 unpaired nucleotides) = ΔH°initiation (3) + ΔH°penalty (C3loops)
Hairpin loops of three nucleotides do not receive a sequence-dependent first mismatch term. All C hairpin loops of three nucleotides receive a stability penalty.
Special hairpin loops
Hairpin loops of 3, 4, and 6 nucleotides that have stabilities poorly fit by the free energy model are assigned enthalpy changes based on experimental data.
5.3 Examples
6 nucleotide hairpin loop with no special stacking terms
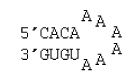
ΔG°37 = ΔG°37(Watson-Crick-Franklin Helix) + ΔG°37(Hairpin Loop)
ΔG°37 = ΔG°37(Watson-Crick-Franklin Helix) + ΔG°37(terminal mismatch) + ΔG°37 Hairpin initiation(6)
ΔG°37 = ΔG°37(CG followed by AU) + ΔG°37(AU followed by CG) + ΔG°37(CG followed by AU) + ΔG°37 AU end penalty + ΔG°37(AU followed by AA) + ΔG°37 Hairpin initiation(6)
ΔG°37 = –2.11 kcal/mol – 2.24 kcal/mol – 2.11 kcal/mol + 0.45 kcal/mol – 0.8 kcal/mol + 5.4 kcal/mol
ΔG°37 = –1.4 kcal/mol
ΔH° = ΔH°(Watson-Crick-Franklin Helix) + ΔH°(Hairpin Loop)
ΔH° = ΔH°(Watson-Crick-Franklin Helix) + ΔH°(terminal mismatch) + ΔH°Hairpin initiation(6)
ΔH° = ΔH°(CG followed by AU) + ΔH°(AU followed by CG) + ΔH°(CG followed by AU) + ΔH°AU end penalty + ΔH°(AU followed by AA) + ΔH°Hairpin initiation(6)
ΔH° = –10.44 kcal/mol – 11.40 kcal/mol – 10.44 kcal/mol + 3.72 kcal/mol – 3.9 kcal/mol – 2.9 kcal/mol
ΔH° = –35.4 kcal/mol
Note that for unimolecular secondary structures, the helical intermolecular initiation does not appear.
5 nucleotide hairpin loop with a GG first mismatch
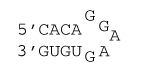
ΔG°37 = ΔG°37(Watson-Crick-Franklin Helix) + ΔG°37(Hairpin Loop)
ΔG°37 = ΔG°37(Watson-Crick-Franklin Helix) + ΔG°37(terminal mismatch) + ΔG°37(GG first mismatch) + ΔG°37 Hairpin initiation(5)
ΔG°37 = ΔG°37(CG followed by AU) + ΔG°37(AU followed by CG) + ΔG°37(CG followed by AU) + ΔG°37 AU end penalty + ΔG°37(AU followed by GG) + ΔG°37(GG first mismatch) + ΔG°37 Hairpin initiation(5)
ΔG°37 = –2.11 kcal/mol – 2.24 kcal/mol – 2.11 kcal/mol + 0.45 kcal/mol – 0.8 kcal/mol – 0.8 kcal/mol + 5.7 kcal/mol
ΔG°37 = –1.9 kcal/mol
ΔH° = ΔH°(Watson-Crick-Franklin Helix) + ΔH°(Hairpin Loop)
ΔH° = ΔH°(Watson-Crick-Franklin Helix) + ΔH°(terminal mismatch) + ΔH°Hairpin initiation(5)
ΔH° = ΔH°(CG followed by AU) + ΔH°(AU followed by CG) + ΔH°(CG followed by AU) + ΔH°AU end penalty + ΔH°(AU followed by GG) + ΔH°Hairpin initiation(5)
ΔH° = –10.44 kcal/mol – 11.40 kcal/mol – 10.44 kcal/mol + 3.72 kcal/mol – 3.5 kcal/mol + 3.6 kcal/mol
ΔH° = –28.5 kcal/mol
4 nucleotide special hairpin loop
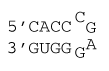
ΔG°37 = ΔG°37(Watson-Crick-Franklin Helix) + ΔG°37(Hairpin Loop)
ΔG°37 = ΔG°37(Watson-Crick-Franklin Helix) + ΔG°37(CcgagG)
ΔG°37 = ΔG°37(CG followed by AU) + ΔG°37(AU followed by CG) + ΔG°37(CG followed by CG) + ΔG°37(CcgagG)
ΔG°37 = –2.11 kcal/mol – 2.24 kcal/mol – 3.26 kcal/mol + 3.5 kcal/mol
ΔG°37 = –4.1 kcal/mol
ΔH° = ΔH°(Watson-Crick-Franklin Helix) + ΔH°(Hairpin Loop)
ΔH° = ΔH°(Watson-Crick-Franklin Helix) + ΔH°(CcgagG)
ΔH° = ΔH°(CG followed by AU) + ΔH°(AU followed by CG) + ΔH°(CG followed by CG) + ΔH°(CcgagG)
ΔH° = –10.44 kcal/mol – 11.40 kcal/mol – 13.39 kcal/mol – 6.6 kcal/mol
ΔH° = –41.8 kcal/mol
6 nucleotide all C loop
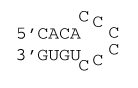
ΔG°37 = ΔG°37(Watson-Crick-Franklin Helix) + ΔG°37(Hairpin Loop)
ΔG°37 = ΔG°37(Watson-Crick-Franklin Helix) + ΔG°37(terminal mismatch) + ΔG°37 Hairpin initiation(6) + ΔG°37 penalty (all C loops)
ΔG°37 = ΔG°37(CG followed by AU) + ΔG°37(AU followed by CG) + ΔG°37(CG followed by AU) + ΔG°37 AU end penalty + ΔG°37(AU followed by CC) + ΔG°37 Hairpin initiation(6) + 6×A + B
ΔG°37 = –2.11 kcal/mol – 2.24 kcal/mol – 2.11 kcal/mol + 0.45 kcal/mol – 0.7 kcal/mol + 5.4 kcal/mol + 6×0.3 kcal/mol + 1.6 kcal/mol
ΔG°37 = 2.1 kcal/mol
ΔH° = ΔH°(Watson-Crick-Franklin Helix) + ΔH°(Hairpin Loop)
ΔH° = ΔH°(Watson-Crick-Franklin Helix) + ΔH°(terminal mismatch) + ΔH°Hairpin initiation(6) + ΔH°penalty (all C loops)
ΔH° = ΔH°(CG followed by AU) + ΔH°(AU followed by CG) + ΔH°(CG followed by AU) + ΔH°AU end penalty + ΔH°(AU followed by CC) + ΔH°Hairpin initiation(6) + 6×A + B
ΔH° = –10.44 kcal/mol – 11.40 kcal/mol – 10.44 kcal/mol + 3.72 kcal/mol + 6.0 kcal/mol – 2.9 kcal/mol + 6×3.4 kcal/mol + 7.6 kcal/mol
ΔH° = +2.5 kcal/mol
5 nucleotide loop with special GU closure
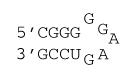
ΔG°37 = ΔG°37(Watson-Crick-Franklin Helix) + ΔG°37(Hairpin Loop)
ΔG°37 = ΔG°37(Watson-Crick-Franklin Helix) + ΔG°37(terminal mismatch) + ΔG°37(GG first mismatch) + ΔG°37 Hairpin initiation(5) + ΔG°37 (special GU closure)
ΔG°37 = ΔG°37(CG followed by GC) + ΔG°37(GC followed by GC) + ΔG°37(GC followed by GU) + ΔG°37 GU end penalty + ΔG°37(GU followed by GG) + ΔG°37(GG first mismatch) + ΔG°37 Hairpin initiation(5) + ΔG°37 (special GU closure)
ΔG°37 = –2.36 kcal/mol – 3.26 kcal/mol – 1.53 kcal/mol + 0.45 kcal/mol – 0.8 kcal/mol – 0.8 kcal/mol + 5.7 kcal/mol – 2.2 kcal/mol
ΔG°37 = –4.8 kcal/mol
ΔH° = ΔH°(Watson-Crick-Franklin Helix) + ΔH°(Hairpin Loop)
ΔH° = ΔH°(Watson-Crick-Franklin Helix) + ΔH°(terminal mismatch) + ΔH°Hairpin initiation(5) + ΔH°(special GU closure)
ΔH° = ΔH°(CG followed by GC) + ΔH°(GC followed by GC) + ΔH°(GC followed by GU) + ΔH°GU end + ΔH°(GU followed by GG) + ΔH°Hairpin initiation(5) + ΔH°(special GU closure)
ΔH° = –10.64 kcal/mol – 13.39 kcal/mol – 8.33 kcal/mol + 3.72 kcal/mol – 3.5 kcal/mol + 3.6 kcal/mol – 14.8 kcal/mol
ΔH° = –43.3 kcal/mol
5.4 Parameter Tables
Length dependent initiation parameters are available in plain text for free energy changes and plain text for enthalpy changes. The plain text initiation tables include an extrapolation out to lengths of 30 nucleotides. These initiation parameters are also available in html format. Initiation parameters are based on experiments for sizes up to 9 nucleotides, but can be extrapolated to longer loops. For free energy changes, the extrapolation is ΔG°37 initiation (n>9) = ΔG°37 initiation (9) + 1.75 RT ln(n/9), where R is the gas constant and T is the absolute temperature. For enthalpy changes, ΔH°initiation (n>9) = ΔH°initiation (9).
The terminal mismatch tables are available in plain text for free energy changes and plain text for enthalpy changes. These parameters are also available in html for free energy changes and html for enthalpy changes.
The bonus/penalty terms (including the all-C loop terms) are available in html format.
The table of special hairpin loops is available in plain text for free energy change for 3, 4, or 6 nucleotides; plain text for enthalpy change for 3, 4, or 6 nucleotides; and in html. The special hairpin loop sequences include the identity of the closing basepair.
5.5 References
The hairpin loop nearest neighbor parameters for free energy change were reported in:
Mathews, D.H., Disney, M.D., Childs, J.L., Schroeder, S.J., Zuker, M. and Turner, D.H. (2004) Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. USA, 101, 7287-7292.
The enthalpy change parameters were reported in:
Lu, Z.J., Turner, D.H. and Mathews, D.H. (2006) A set of nearest neighbor parameters for predicting the enthalpy change of RNA secondary structure formation. Nucleic Acids Res., 34 4912 - 4924.
The experimental data for the fit of the parameters were taken from:
- Groebe, D.R. and Uhlenbeck, O.C. (1988) Characterization of RNA hairpin loop stability. Nucleic Acids Res., 16, 11725-11735.
- Antao, V.P., Lai, S.Y. and Tinoco, I., Jr. (1991) A thermodynamic study of unusually stable RNA and DNA hairpins. Nucleic Acids Res., 19, 5901-5905.
- Antao, V.P. and Tinoco, I., Jr. (1992) Thermodynamic parameters for loop formation in RNA and DNA hairpin tetraloops. Nucleic Acids Res., 20, 819-824.
- Serra, M.J., Lyttle, M.H., Axenson, T.J., Schadt, C.A. and Turner, D.H. (1993) RNA hairpin loop stability depends on closing pair. Nucleic Acids Res., 21, 3845-3849.
- Serra, M.J., Axenson, T.J. and Turner, D.H. (1994) A model for the stabilities of RNA hairpins based on a study of the sequence dependence of stability for hairpins of six nucleotides. Biochemistry, 33, 14289-14296.
- Laing, L.G. and Hall, K.B. (1996) A model of the iron responsive element RNA hairpin loop structure determined from NMR and thermodynamic data. Biochemistry, 35, 13586-13596.
- Serra, M.J., Barnes, T.W., Betschart, K., Gutierrez, M.J., Sprouse, K.J., Riley, C.K., Stewart, L. and Temel, R.E. (1997) Improved parameters for the prediction of RNA hairpin stability. Biochemistry, 36, 4844-4851.
- Giese, M.R., Betschart, K., Dale, T., Riley, C.K., Rowan, C., Sprouse, K.J. and Serra, M.J. (1998) Stability of RNA hairpins closed by wobble base pairs. Biochemistry, 37, 1094-1100.
- Shu, Z. and Bevilacqua, P.C. (1999) Isolation and characterization of thermodynamically stable and unstable RNA hairpins from a triloop combinatorial library. Biochemistry, 38, 15369-15379.
- Dale, T., Smith, R. and Serra, M. (2000) A test of the model to predict unusually stable RNA hairpin loop stability. RNA, 6, 608-615.
- Proctor, D.J., Schaak, J.E., Bevilacqua, J.M., Falzone, C.J. and Bevilacqua, P.C. (2002) Isolation and characterization of stable tetraloops with the motif YNMG that participates in tertiary interactions. Biochemistry, 41, 12062-12075.
