19 Multibranch Loops
19.1 Folding Free Energy Change
Multibranch loops stabilities are predicted using the following equation:
ΔG°37 multibranch = ΔG°37 initiation + ΔG°37 stacking
where the stacking is the optimal configuration of dangling ends, terminal mismatches, or coaxial stacks, noting that each nucleotide or helix end can participate in only one of these favorable interactions.
When there are six or fewer unpaired nucleotides in the loop, initiation is predicted using:
ΔG°37 initiation = a + b×[number of unpaired nucleotides] + c×[number of branching helices]
where a, b, and c are parameters.
For multibranch loops with greater than six unpaired nucleotides, a logarithmic dependence on the number of upaired nucleotides is used:
ΔG°37 initiation = a + 6b + (1.1 kcal/mol)×ln([number of unpaired nucleotides]/6) + c×[number of branching helices]
19.2 Example
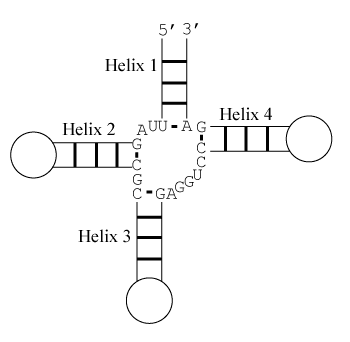
Free Energy Change
Prediction of Stacking
The predicted stacking configuration is the one with lowest free energy change. There are eight relevant configurations.
Configuration 1:
Helix 1 with \(3'\) dangling U, Helix 2 with terminal mismatch, Helix 3 with \(3'\) dangling A, and Helix 4 with \(5'\) dangling C
ΔG°37 = ΔG°37(UA with \(3'\) dangling U) + ΔG°37(CG followed by GA) + ΔG°37(GC with \(3'\) dangling A) + ΔG°37(GC with \(5'\) dangling C)
ΔG°37 = –0.1 kcal/mol – 1.4 kcal/mol – 1.1 kcal/mol – 0.3 kcal/mol
ΔG°37 = –2.9 kcal/mol
Configuration 2:
Helix 1 with \(3'\) dangling U, Helix 2 with \(5'\) dangling A, Helix 3 with terminal mismatch, and Helix 4 with \(5'\) dangling C
ΔG°37 = ΔG°37(UA with \(3'\) dangling U) + ΔG°37(CG with \(5'\) dangling A) + ΔG°37(GC followed by AG) + ΔG°37(GC with \(5'\) dangling C)
ΔG°37 = –0.1 kcal/mol – 0.2 kcal/mol – 1.3 kcal/mol – 0.3 kcal/mol
ΔG°37 = –1.9 kcal/mol
Configuration 3:
Helix 1 in flush coaxial stack with helix 4, Helix 2 with terminal mismatch, and Helix 3 with \(3'\) dangling A
ΔG°37 = ΔG°37(GC followed by AU) + ΔG°37(CG followed by GA) + ΔG°37(GC with \(3'\) dangling A)
ΔG°37 = –2.35 kcal/mol – 1.4 kcal/mol – 1.1 kcal/mol
ΔG°37 = –4.9 kcal/mol
Configuration 4:
Helix 1 in flush coaxial stack with helix 4, Helix 2 with \(5'\) dangling A, and Helix 3 with terminal mismatch
ΔG°37 = ΔG°37(GC followed by AU) + ΔG°37(CG with \(5'\) dangling A) + ΔG°37(GC followed by AG)
ΔG°37 = –2.35 kcal/mol – 0.2 kcal/mol – 1.3 kcal/mol
ΔG°37 = –3.9 kcal/mol
Configuration 5:
Helix 1 with \(3'\) dangling U, Helix 2 in mismatch–mediated coaxial stack with helix 3 with GA intervening mismatch, and Helix 4 with \(5'\) dangling C
ΔG°37 = ΔG°37(UA with \(3'\) dangling U) + ΔG°37(CG followed by GA) + ΔG°37(Discontinuous Backbone Stack) + ΔG°37(GC with \(5'\) dangling C)
ΔG°37 = –0.1 kcal/mol – 1.4 kcal/mol – 2.1 kcal/mol – 0.3 kcal/mol
ΔG°37 = –3.9 kcal/mol
Configuration 6:
Helix 1 with \(3'\) dangling U, Helix 2 in mismatch–mediated coaxial stack with helix 3 with AG intervening mismatch, and Helix 4 with \(5'\) dangling C
ΔG°37 = ΔG°37(UA with \(3'\) dangling U) + ΔG°37(Discontinuous Backbone Stack) + ΔG°37(GC followed by AG) + ΔG°37(GC with \(5'\) dangling C)
ΔG°37 = –0.1 kcal/mol – 2.1 kcal/mol – 1.3 kcal/mol – 0.3 kcal/mol
ΔG°37 = –3.8 kcal/mol
Configuration 7:
Helix 1 in flush coaxial stack with helix 4 and Helix 2 in mismatch–mediated coaxial stack with helix 3 with GA intervening mismatch
ΔG°37 = ΔG°37(GC followed by AU) + ΔG°37(CG followed by GA) + ΔG°37(Discontinuous Backbone Stack)
ΔG°37 = –2.35 kcal/mol – 1.4 kcal/mol – 2.1 kcal/mol
ΔG°37 = –5.9 kcal/mol
Configuration 8:
Helix 1 in flush coaxial stack with helix 4 and Helix 2 in mismatch–mediated coaxial stack with helix 3 with AG intervening mismatch
ΔG°37 = ΔG°37(GC followed by AU) + ΔG°37(Discontinuous Backbone Stack) + ΔG°37(GC followed by AG)
ΔG°37 = –2.35 kcal/mol – 2.1 kcal/mol – 1.3 kcal/mol
ΔG°37 = –5.8 kcal/mol
Configuration 7 has the lowest folding free energy change of –5.9 kcal/mol.
Initiation Free Energy Change
ΔG°37 initiation = a + 6b + (1.1 kcal/mol)×ln([number of unpaired nucleotides]/6) + c×[number of branching helices]
ΔG°37 initiation = 10.1 kcal/mol + 6×(–0.3 kcal/mol) + (1.1 kcal/mol)×ln(8/6) + 4×(–0.3 kcal/mol)
ΔG°37 initiation = 7.4 kcal/mol
19.2.1 Total Folding Free Energy Change
ΔG°37 multibranch loop = ΔG°37 initiation + ΔG°37 stacking = 7.4 kcal/mol – 5.9 kcal/mol = 1.5 kcal/mol
Note that helices 1 and 2 are separated by two unpaired nucleotides and cannot stack coaxially. Similarly, helices 3 and 4 are too distant to stack coaxially. Also note that coaxial stacking is only allowed between adjacent helices and hence, for example, helices 1 and 3 cannot stack coaxially.
19.3 Tables
Tables of parameters are available in html format.
19.4 References
The multibranch loop nearest neighbor parameters for initiation free energy change were reported in:
Mathews, D.H., Sabina, J., Zuker, M. and Turner, D.H. (1999) Expanded sequence dependence of thermodynamic parameters provides improved prediction of RNA secondary structure. J. Mol. Biol., 288, 911-940.
