4 Terminal Mismatches
Terminal mismatches are non-canonical pairs adjacent to helix ends.
4.1 Example
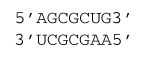
ΔG°37 = ΔG°37(Watson-Crick-Franklin Helix) + ΔG°37(UA followed by GA)
ΔG°37 = –7.94 kcal/mol – 1.1 kcal/mol
ΔG°37 = –9.0 kcal/mol
ΔH° = ΔH°(Watson-Crick-Franklin Helix) + ΔH°(UA followed by GA)
ΔH° = –50.31 kcal/mol – 3.8 kcal/mol
ΔH° = –54.1 kcal/mol
4.2 Tables
Tables of parameters are available as plain text for free energy changes, plain text for enthalpy changes, html for free energy change, or html for enthalpy change.
4.3 References
The terminal mismatch parameters were assembled in:
Xia, T., Mathews, D.H. and Turner, D.H. (1999) In Söll, D. G., Nishimura, S. and Moore, P. B. (eds.), Prebiotic Chemistry, Molecular Fossils, Nucleosides, and RNA. Elsevier, New York, pp. 21-47.
The optical melting experiments for terminal mismatches were reported in:
- Hickey, D.R. and Turner, D.H. (1985) Solvent effects on the stability of A7U7p. Biochemistry, 24, 2086-2094.
- Freier, S.M., Kierzek, R., Caruthers, M.H., Neilson, T. and Turner, D.H. (1986) Free energy contributions of G.U and other terminal mismatches to helix stability. Biochemistry, 25, 3209-3223.
- Freier, S.M., Sugimoto, N., Sinclair, A., Alkema, D., Neilson, T., Kierzek, R., Caruthers, M.H. and Turner, D.H. (1986) Stability of XGCGCp, GCGCYp, and XGCGCYp helixes: an empirical estimate of the energetics of hydrogen bonds in nucleic acids. Biochemistry, 25, 3214-3219.
- Sugimoto, N., Kierzek, R. and Turner, D.H. (1987) Sequence dependence for the energetics of dangling ends and terminal base pairs in ribonucleic acid. Biochemistry, 26, 4554-4558.
- Serra, M.J., Axenson, T.J. and Turner, D.H. (1994) A model for the stabilities of RNA hairpins based on a study of the sequence dependence of stability for hairpins of six nucleotides. Biochemistry, 33, 14289-14296.
- Dale, T., Smith, R. and Serra, M. (2000) A test of the model to predict unusually stable RNA hairpin loop stability. RNA, 6, 608-615.
