14 Terminal Mismatches
Terminal mismatches are non-canonical pairs adjacent to helix ends.
14.1 Example
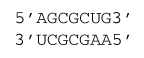
ΔG°37 = ΔG°37(Watson-Crick-Franklin Helix) + ΔG°37(UA followed by GA)
ΔG°37 = –7.94 kcal/mol – 1.1 kcal/mol
ΔG°37 = –9.0 kcal/mol
14.2 Tables
14.3 References
The derivation of terminal mismatch parameters was described in:
Mathews, D.H., Sabina, J., Zuker, M. and Turner, D.H. (1999) Expanded sequence dependence of thermodynamic parameters provides improved prediction of RNA secondary structure. J. Mol. Biol., 288, 911-940.
The optical melting experiments for terminal mismatches were reported in:
- Hickey, D.R. and Turner, D.H. (1985) Solvent effects on the stability of A7U7p. Biochemistry, 24, 2086-2094.
- Freier, S.M., Kierzek, R., Caruthers, M.H., Neilson, T. and Turner, D.H. (1986) Free energy contributions of G.U and other terminal mismatches to helix stability. Biochemistry, 25, 3209-3223.
- Freier, S.M., Sugimoto, N., Sinclair, A., Alkema, D., Neilson, T., Kierzek, R., Caruthers, M.H. and Turner, D.H. (1986) Stability of XGCGCp, GCGCYp, and XGCGCYp helixes: an empirical estimate of the energetics of hydrogen bonds in nucleic acids. Biochemistry, 25, 3214-3219.
- Sugimoto, N., Kierzek, R. and Turner, D.H. (1987) Sequence dependence for the energetics of dangling ends and terminal base pairs in ribonucleic acid. Biochemistry, 26, 4554-4558.
- Serra, M.J., Axenson, T.J. and Turner, D.H. (1994) A model for the stabilities of RNA hairpins based on a study of the sequence dependence of stability for hairpins of six nucleotides. Biochemistry, 33, 14289-14296.
- Dale, T., Smith, R. and Serra, M. (2000) A test of the model to predict unusually stable RNA hairpin loop stability. RNA, 6, 608-615.
