2 GU Pairs
GU pairs are generally treated as nearest neighbor stacks, similar to Watson-Crick-Franklin helices, and GU pairs at the ends of helices are penalized with the same parameter as AU pairs at the ends of helices. In one sequence context, a tandem GU pair with a GU followed by a UG, the nearest neighbor model does not work and two parameters are available, depending on the sequence context (see the html table of parameters). Note also that the motif \(5'\)GG/\(3'\)UU was assigned a ΔG°37 of –0.5 kcal/mol to optimize structure prediction accurracy, whereas it was measured as +0.5 kcal/mol. Parameters for stacks containing GU pairs were calculated separately from those containing AU and GC base pairs only.
2.1 Example
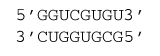
ΔG°37 = ΔG°37 intermolecular initiation + ΔG°37 GU end penalty + ΔG°37(GC followed by GU, followed by UG, followed by CG) + ΔG°37(CG followed by GU) + ΔG°37(GU followed by UG) + ΔG°37(UG followed by GC) + ΔG°37(GC followed by UG)
ΔG°37 = 4.09 kcal/mol + 0.45 kcal/mol – 4.12 kcal/mol –1.41 kcal/mol + 1.29 kcal/mol – 1.41 kcal/mol – 2.51 kcal/mol
ΔG°37 = –3.62 kcal/mol
ΔH° = ΔH°intermolecular initiation + ΔH°GU end penalty + ΔH°(GC followed by GU, followed by UG, followed by GC) + ΔH°(CG followed by GU) + ΔH°(GU followed by UG) + ΔH°(UG followed by GC) + ΔH°(GC followed by UG)
ΔH° = 3.61 kcal/mol + 3.72 kcal/mol – 30.80 kcal/mol – 5.61 kcal/mol – 14.59 kcal/mol – 5.61 kcal/mol – 12.59 kcal/mol
ΔH° = –61.87 kcal/mol
Note that this example shows the stack of GU followed by UG in two different contexts, including the stabilizing context and the destabilizing context. In the stabilizing context, a single parameter is used for three consecutive basepair stacks.
2.2 Tables
The tables of parameters are available as plain text for free energy change (including Watson-Crick-Franklin pairs), plain text for enthalpy change (including Watson-Crick-Franklin pairs), or html.
2.3 References
The GU nearest neighbor parameters were reported in:
Mathews, D.H., Sabina, J., Zuker, M. and Turner, D.H. (1999) Expanded sequence dependence of thermodynamic parameters provides improved prediction of RNA secondary structure. J. Mol. Biol., 288, 911-940.
The experimental data for the fit of the parameters were taken from:
- Freier, S.M., Kierzek, R., Caruthers, M.H., Neilson, T. and Turner, D.H. (1986) Free energy contributions of G.U and other terminal mismatches to helix stability. Biochemistry, 25, 3209-3223.
- Sugimoto, N., Kierzek, R., Freier, S.M. and Turner, D.H. (1986) Energetics of internal GU mismatches in ribooligonucleotide helixes. Biochemistry, 25, 5755-5759.
- He, L., Kierzek, R., SantaLucia, J., Jr., Walter, A.E. and Turner, D.H. (1991) Nearest-neighbor parameters for G.U mismatches. Biochemistry, 30, 11124-11132.
- Wu, M., McDowell, J.A. and Turner, D.H. (1995) A periodic table of symmetric tandem mismatches in RNA. Biochemistry, 34, 3204-3211.
- McDowell, J.A. and Turner, D.H. (1996) Investigation of the structural basis for thermodynamic stabilities of tandem GU mismatches: Solution structure of (rGAGGUCUC)2 by two-dimensional NMR and simulated annealing. Biochemistry, 35, 14077-14089.
- Xia, T., McDowell, J.A. and Turner, D.H. (1997) Thermodynamics of nonsymmetric tandem mismatches adjacent to G.C base pairs in RNA. Biochemistry, 36, 12486-12487.
