RNA is an important type of biological molecule. It is used by cells to express genes and to regulate gene expression. It can catalyze reactions. RNA is also an important drug target. The Mathews lab studies RNA using computational methods.
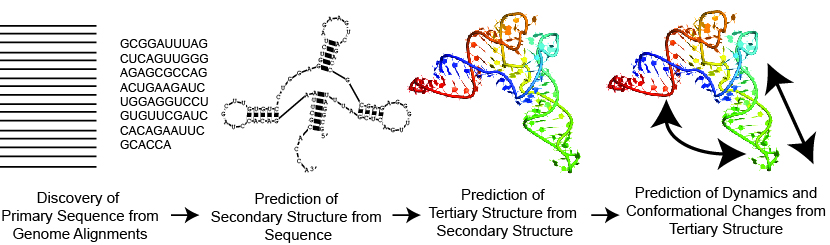
Our goal is to automate the modeling of RNA structure and function from genome sequence to 3D structure (as shown in the Figure above). This is important because it improves our understanding of Biology and also because it helps us cure diseases. We develop software tools to predict the structure of RNA, to model conformational flexibility, to find structured genes and genomes, and to design RNA sequences that will fold into specific structures. Our RNAstructure package predicts and analyzes RNA secondary structure, i.e. the set of canonical base pairs that the RNA forms. We use and improve Amber for modeling of three-dimensional structures.
We are currently funded with grants from the National Institutes of Health.