Free Energy Change
Prediction of Stacking
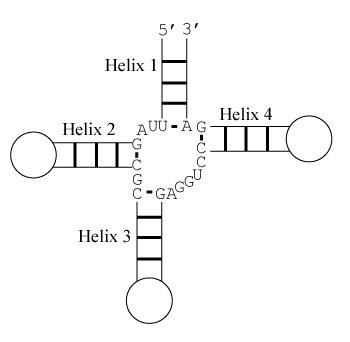
The predicted stacking configuration is the one with lowest free energy change. There are eight relevant configurations.
Configuration 1:
Helix 1 with 3' dangling U, Helix 2 with terminal mismatch, Helix 3 with 3' dangling A, and Helix 4 with 5' dangling C
∆G°37 = ∆G°37(UA with 3' dangling U) + ∆G°37(CG followed by GA) + ∆G°37(GC with 3' dangling A) + ∆G°37(GC with 5' dangling C)
∆G°37 = –0.1 kcal/mol – 1.4 kcal/mol – 1.1 kcal/mol – 0.3 kcal/mol
∆G°37 = –2.9 kcal/mol
Configuration 2:
Helix 1 with 3' dangling U, Helix 2 with 5' dangling A, Helix 3 with terminal mismatch, and Helix 4 with 5' dangling C
∆G°37 = ∆G°37(UA with 3' dangling U) + ∆G°37(CG with 5' dangling A) + ∆G°37(GC followed by AG) + ∆G°37(GC with 5' dangling C)
∆G°37 = –0.1 kcal/mol – 0.2 kcal/mol – 1.3 kcal/mol – 0.3 kcal/mol
∆G°37 = –1.9 kcal/mol
Configuration 3:
Helix 1 in flush coaxial stack with helix 4, Helix 2 with terminal mismatch, and Helix 3 with 3' dangling A
∆G°37 = ∆G°37(GC followed by AU) + ∆G°37(CG followed by GA) + ∆G°37(GC with 3' dangling A)
∆G°37 = –2.35 kcal/mol – 1.4 kcal/mol – 1.1 kcal/mol
∆G°37 = –4.9 kcal/mol
Configuration 4:
Helix 1 in flush coaxial stack with helix 4, Helix 2 with 5' dangling A, and Helix 3 with terminal mismatch
∆G°37 = ∆G°37(GC followed by AU) + ∆G°37(CG with 5' dangling A) + ∆G°37(GC followed by AG)
∆G°37 = –2.35 kcal/mol – 0.2 kcal/mol – 1.3 kcal/mol
∆G°37 = –3.9 kcal/mol
Configuration 5:
Helix 1 with 3' dangling U, Helix 2 in mismatch–mediated coaxial stack with helix 3 with GA intervening mismatch, and Helix 4 with 5' dangling C
∆G°37 = ∆G°37(UA with 3' dangling U) + ∆G°37(CG followed by GA) + ∆G°37(Discontinuous Backbone Stack) + ∆G°37(GC with 5' dangling C)
∆G°37 = –0.1 kcal/mol – 1.4 kcal/mol – 2.1 kcal/mol – 0.3 kcal/mol
∆G°37 = –3.9 kcal/mol
Configuration 6:
Helix 1 with 3' dangling U, Helix 2 in mismatch–mediated coaxial stack with helix 3 with AG intervening mismatch, and Helix 4 with 5' dangling C
∆G°37 = ∆G°37(UA with 3' dangling U) + ∆G°37(Discontinuous Backbone Stack) + ∆G°37(GC followed by AG) + ∆G°37(GC with 5' dangling C)
∆G°37 = –0.1 kcal/mol – 2.1 kcal/mol – 1.3 kcal/mol – 0.3 kcal/mol
∆G°37 = –3.8 kcal/mol
Configuration 7:
Helix 1 in flush coaxial stack with helix 4 and Helix 2 in mismatch–mediated coaxial stack with helix 3 with GA intervening mismatch
∆G°37 = ∆G°37(GC followed by AU)) + ∆G°37(CG followed by GA) + ∆G°37(Discontinuous Backbone Stack)
∆G°37 = –2.35 kcal/mol – 1.4 kcal/mol – 2.1 kcal/mol
∆G°37 = –5.9 kcal/mol
Configuration 8:
Helix 1 in flush coaxial stack with helix 4 and Helix 2 in mismatch–mediated coaxial stack with helix 3 with AG intervening mismatch
∆G°37 = ∆G°37(GC followed by AU)) + ∆G°37(Discontinuous Backbone Stack) + ∆G°37(GC followed by AG)
∆G°37 = –2.35 kcal/mol – 2.1 kcal/mol – 1.3 kcal/mol
∆G°37 = –5.8 kcal/mol
Configuration 7 has the lowest folding free energy change of –5.9 kcal/mol.
Initiation Free Energy Change
ΔG°37 initiation = a + b×[average asymmetry] + c×[number of branching helices] + ΔG°37 strain(three–way branching loops with fewer than two unpaired nucleotides)
ΔG°37 initiation = 9.25 kcal/mol + (0.91 kcal/mol)×[average asymmetry] + (–0.63 kcal/mol)×[4]
Average asymmetry = min[2.0,(2+1+4+5)/4] = min[2.0,3.0] = 2.0
ΔG°37 initiation = 9.25 kcal/mol + (0.91 kcal/mol)×[2] + (–0.63 kcal/mol)×[4]
ΔG°37 initiation = 8.6 kcal/mol
Total Folding Free Energy Change
ΔG°37 multibranch loop = ΔG°37 initiation + ΔG°37 stacking = 8.6 kcal/mol – 5.9 kcal/mol = 3.6 kcal/mol
Enthalpy Change
Prediction of Stacking
The stacking configuration is fixed by the prediction of folding free energy change and is configuration 7 above.
∆H°= ∆H°(GC followed by AU)) + ∆H°(CG followed by GA) + ∆H°(Discontinuous Backbone Stack)
∆H° = –12.44 kcal/mol – 8.2 kcal/mol – 8.46 kcal/mol
∆H° = –29.1 kcal/mol
Initiation Enthalpy Change
ΔH°initiation = a + b×[average asymmetry] + c×[number of branching helices] + ΔH°strain(three–way branching loops with fewer than two unpaired nucleotides)
ΔH°initiation = 38.9 kcal/mol + (12.9 kcal/mol)×[average asymmetry] + (–11.9 kcal/mol)×[4]
Average asymmetry = min[2.0,(3+2+4+5)/4] = min[2.0,3.0] = 2.0
ΔH°initiation = 38.9 kcal/mol + (12.9 kcal/mol)×[2] + (–11.9 kcal/mol)×[4]
ΔH°initiation = 17.1 kcal/mol
Total Folding Enthalpy Energy Change
ΔH°multibranch loop = ΔH°initiation + ΔH°stacking = 17.1 kcal/mol – 29.1 kcal/mol = –12.0 kcal/mol
Note that helices 1 and 2 are separated by two unpaired nucleotides and cannot stack coaxially. Similarly, helices 3 and 4 are too distant to stack coaxially. Also note that coaxial stacking is only allowed between adjacent helices and hence, for example, helices 1 and 3 cannot stack coaxially.
 Turner 2004
Turner 2004